
Probing the dark matter of regulatory DNA - Though > 90% of disease-causing variants in the human genome occur outside protein-coding regions, how these non-coding variants impact levels of gene expression remains largely unknown. To address this gap, we are leveraging the MITOMI platform to probe how sequence context outside transcription factor binding sites tunes binding kinetics and thermodynamics.
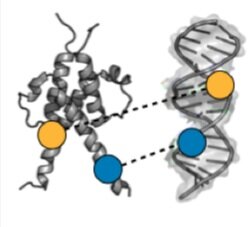
Dissecting the functional effects of transcription factor mutations - Transcription Factor (TF) proteins bind regulatory DNA sequences to control the expression of downstream genes, and mutations to TFs can affect the affinity and specificity of DNA binding. We are using microfluidic devices to quantitatively measure DNA binding affinities and dissociation kinetics for 1000s of TF-DNA interactions in parallel. This approach provides critical experimental data needed to build computational models of effects of TF mutations required for precision medicine.
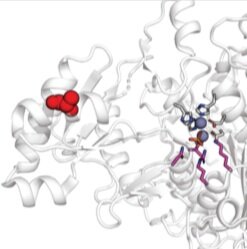
HT-MEK: a new kind of enzymology - We have developed a new platform (HT-MEK, for high-throughput microfluidic enzyme kinetics) that enables, for the first time, the rapid and quantitative measurement of multiple parameters for designed enzyme libraries. To date, we have characterized over 1,100 enzyme mutants (measuring >10,000 individual kinetic and thermodynamic parameters), and have identified structurally segregated regions controlling distinct biochemical properties within a model system phosphatase. Ultimately, these measurements will help yield next-generation predictive models of mutational effects required for rational enzyme design and precision medicine.
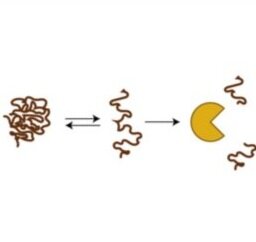
High-throughput and quantitative measurements of protein stability - Disease-associated protein mutations often act by destabilizing protein structure. In personalized medicine, the majority of disease-associated mutations are of unknown significance, preventing clinical action. We are developing parallel assays to quantify folding stability for thousands of proteins variants in parallel, using next-generation sequencing and fluorescence microscopy. With these approaches, we aim to characterize the protein stability landscape and better understand mechanisms of loss-of-function involved in disease.
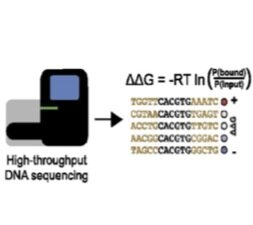
BET-seq: ultra-high-throughput measurement of transcription factor-DNA binding affinities - The rapidly decreasing costs of next-generation sequencing (NGS) present an opportunity to sequence and identify millions of DNA molecules in parallel. Here, we paired MITOMI devices with NGS to quantitatively measure transcription factor binding energies for a million sequences in a single experiment. We are currently applying this platform to study how flanking nucleotides outside the canonical binding site can impact TF-DNA binding affinity.