MRBLE-path - In sepsis, the time to antibiotic treatment is critical for patient survival, and the choice of treatment depends on the pathogen responsible for infection. We are developing a new method to rapidly screen for the presence of multiple bacteria species in a single reaction using oligonucleotide hybridization probes conjugated to MRBLEs with a 1:1 relationship between probe sequence and embedded code, allowing multiplexed measurement of pathogens via imaging alone.
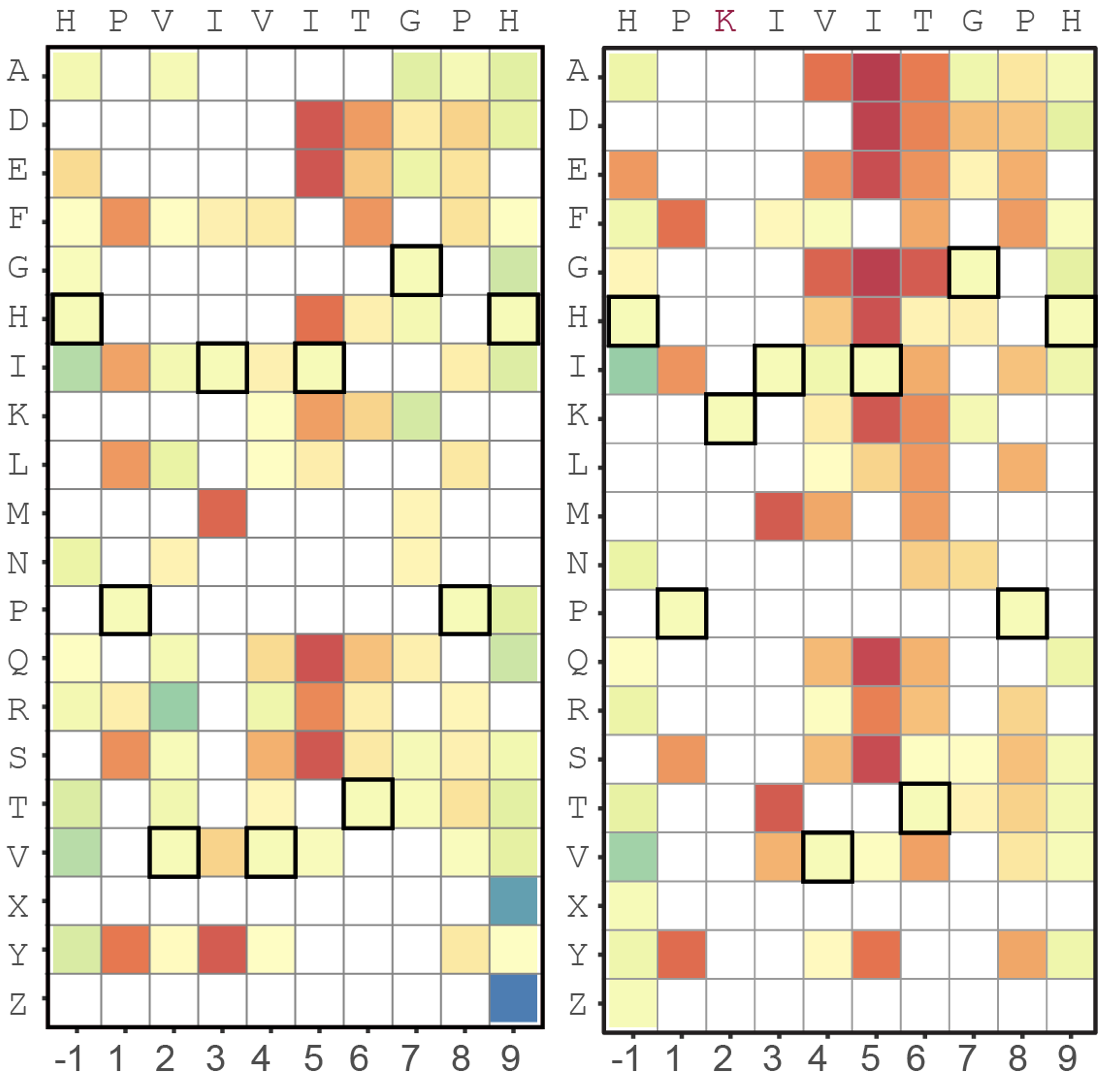
MRBLE-pep - While advances in deep sequencing have dramatically improved our ability to profile how DNA and RNA binding proteins interact with their substrates, we lack similar tools for quantitative, high-throughput investigation of protein-peptide interactions. Here, we synthesize libraries of peptides directly on MRBLEs with a 1:1 linkage between embedded code and peptide sequence, allowing high-throughput measurement of 100 weak protein-peptide interactions in parallel using very little material. In collaboration with the Cyert lab, we are using this platform to understand how human signaling proteins recognize their downstream targets.
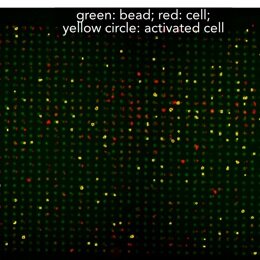
BATTLES - T cells can discriminate between self- and non-self-antigens with exquisite sensitivity and specificity: even a single non-self antigen in a sea of 100,000 self antigens. Despite the central importance of this process for immune surveillance, we do not yet understand the mechanisms involved or how to manipulate them to generate new therapies. We are using MRBLEs to allow high-throughput measurements of how T cell activation depends on presented antigen sequence and applied forces.